Estimating growth rates from age-at-size data
Source:vignettes/growthEstimation.Rmd
growthEstimation.RmdI still need to write this vignette.
library(growthEstimation)
pars <- list(
k = 0.1,
L_inf = 110,
d = 1,
m = 1,
annuli_date = 0,
annuli_min_age = 0,
spawning_mu = 0.4,
spawning_kappa = 10
)
age_at_length <- Cod_CS_age_at_length
plotAgeLikelihood(pars, age_at_length)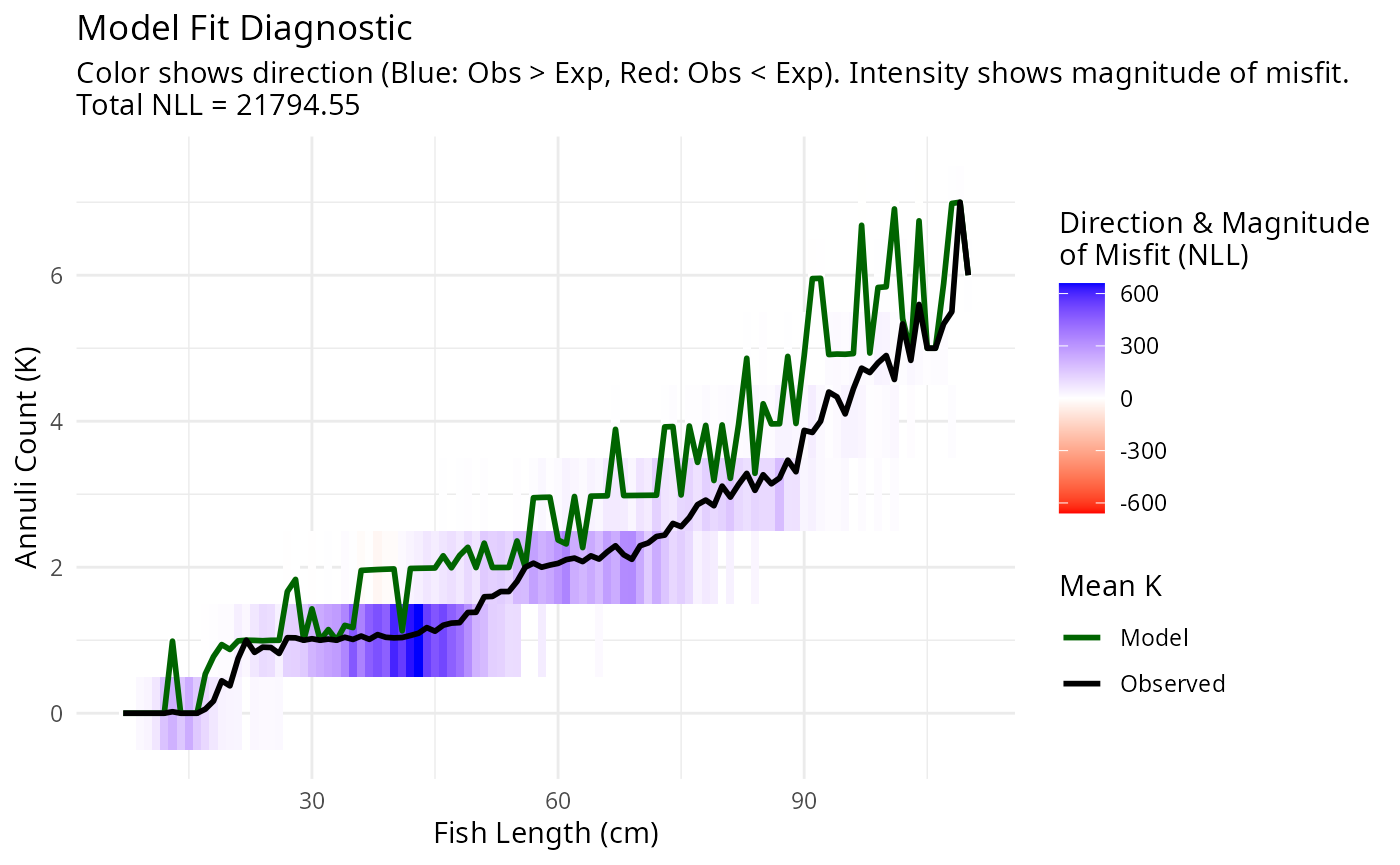
To start tuning parameters, run the following code. A Shiny app will open
tune_pars(pars, age_at_length)You can automatically tune the parameters with
fit <- fit_tmb_nll(pars, surveys = age_at_length)
pars <- fit$pars
plotAgeLikelihood(pars, age_at_length)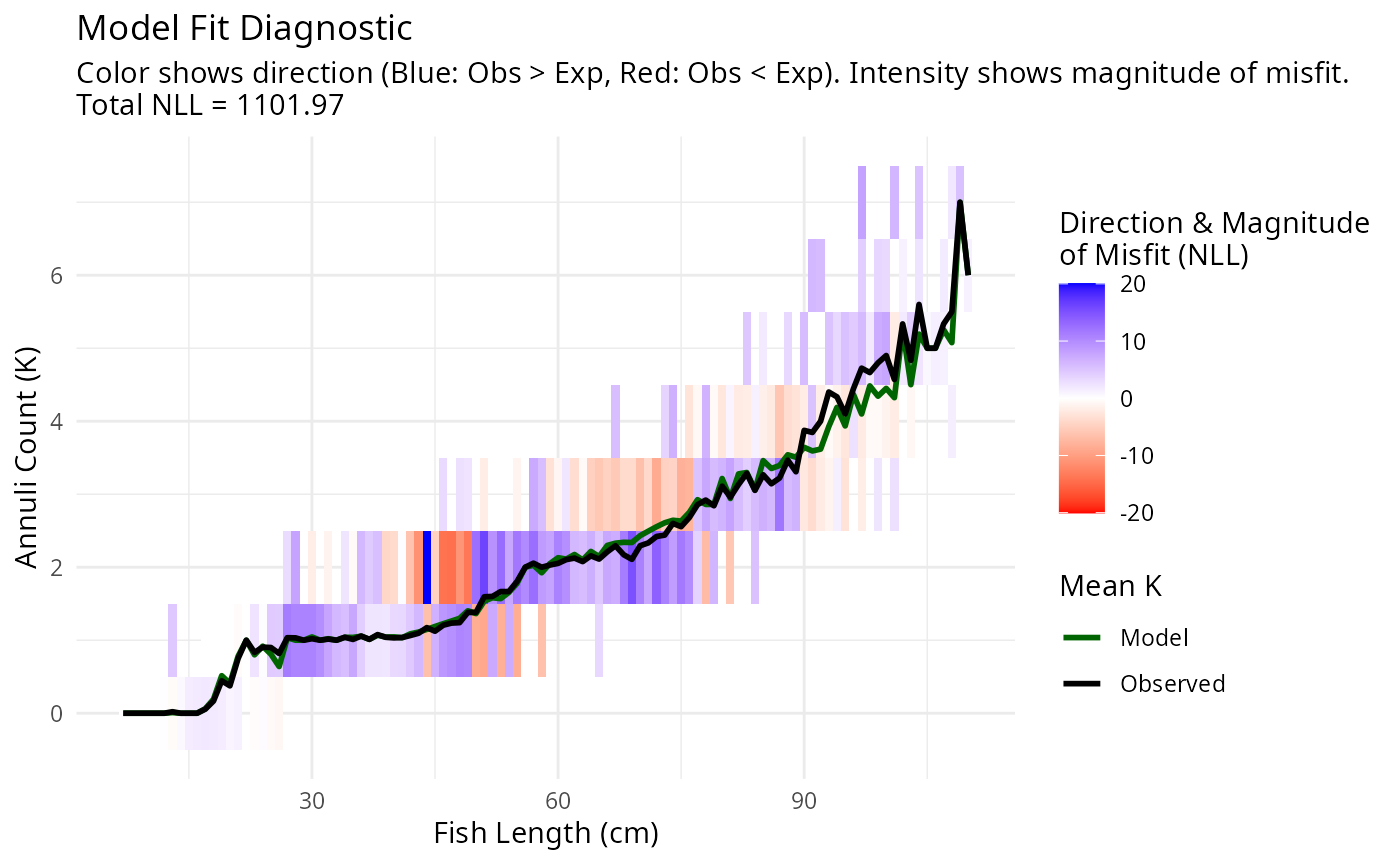
The parameters appear to fit the age-at-size data, but the mortality and diffusion coefficients are too low. The estimated parameters are:
parameter_names <- c("k", "L_inf", "d", "m", "annuli_min_age")
pars[parameter_names]## $k
## [1] 0.1974143
##
## $L_inf
## [1] 164.8323
##
## $d
## [1] 1e-06
##
## $m
## [1] 1e-06
##
## $annuli_min_age
## [1] 0This leads to the following time evolution of a yearly cohort:
u <- getNumberDensity(pars, l_max = 200, t_max = 15)
plotDensity2D(u)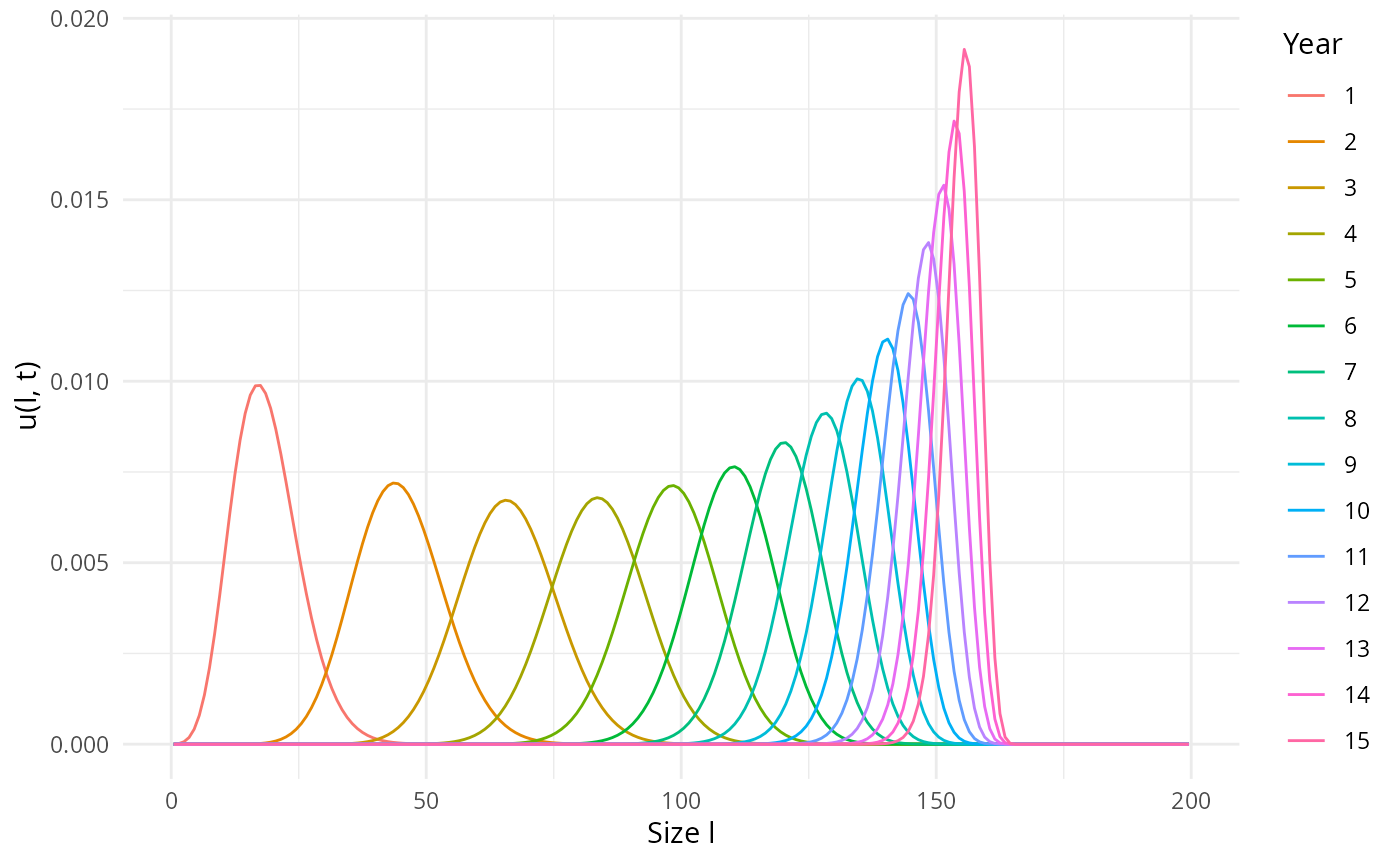
If we constrain the mortality coefficient to be at least 30 we get:
fit <- fit_tmb_nll(pars, surveys = age_at_length,
lower = c(m = 25))
pars <- fit$pars
plotAgeLikelihood(pars, age_at_length)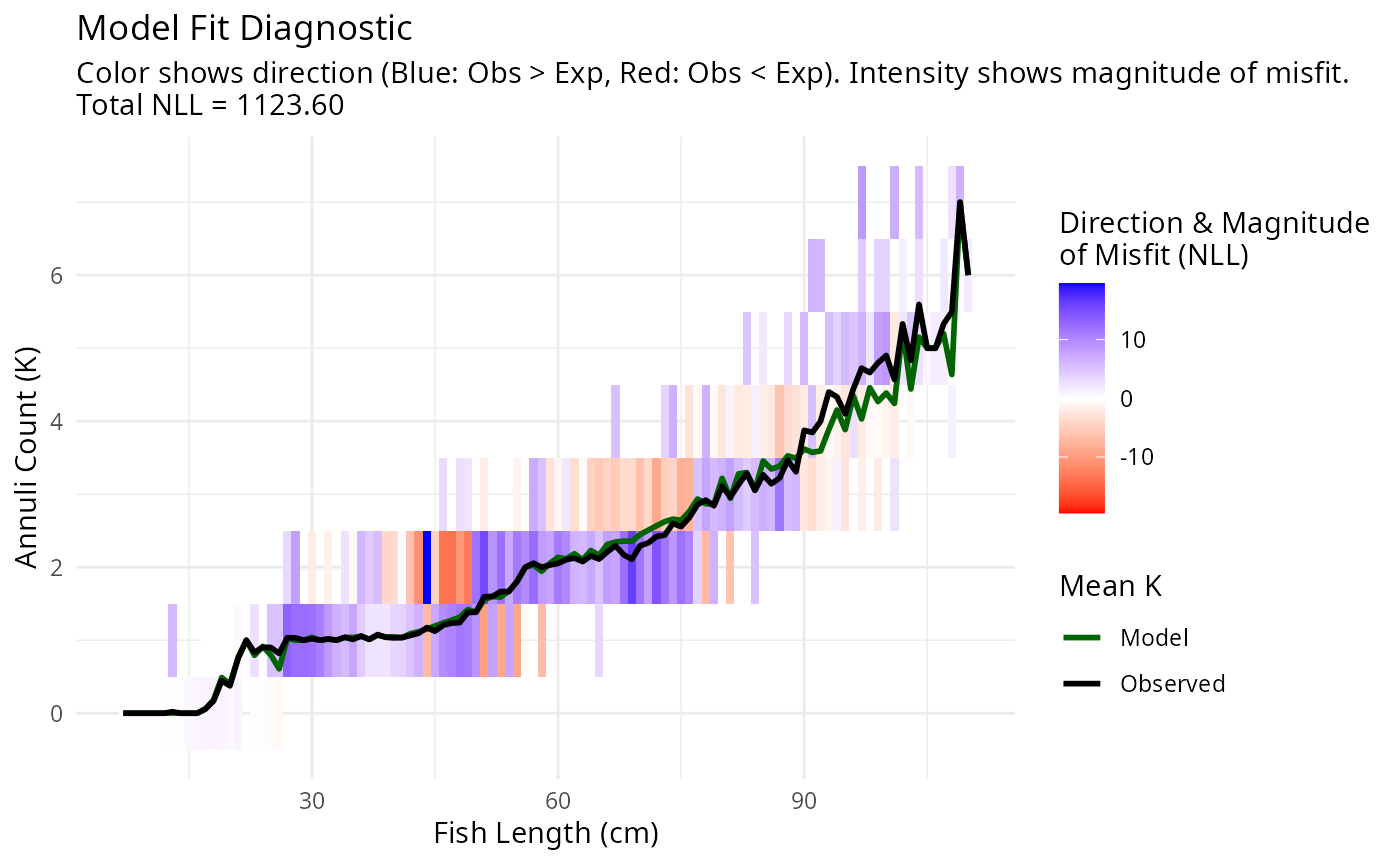
pars[parameter_names]## $k
## [1] 0.1512644
##
## $L_inf
## [1] 192.9295
##
## $d
## [1] 1e-06
##
## $m
## [1] 25
##
## $annuli_min_age
## [1] 0This leads to a more realistic time evolution of a yearly cohort:
u <- getNumberDensity(pars, l_max = 200, t_max = 15)
plotDensity2D(u)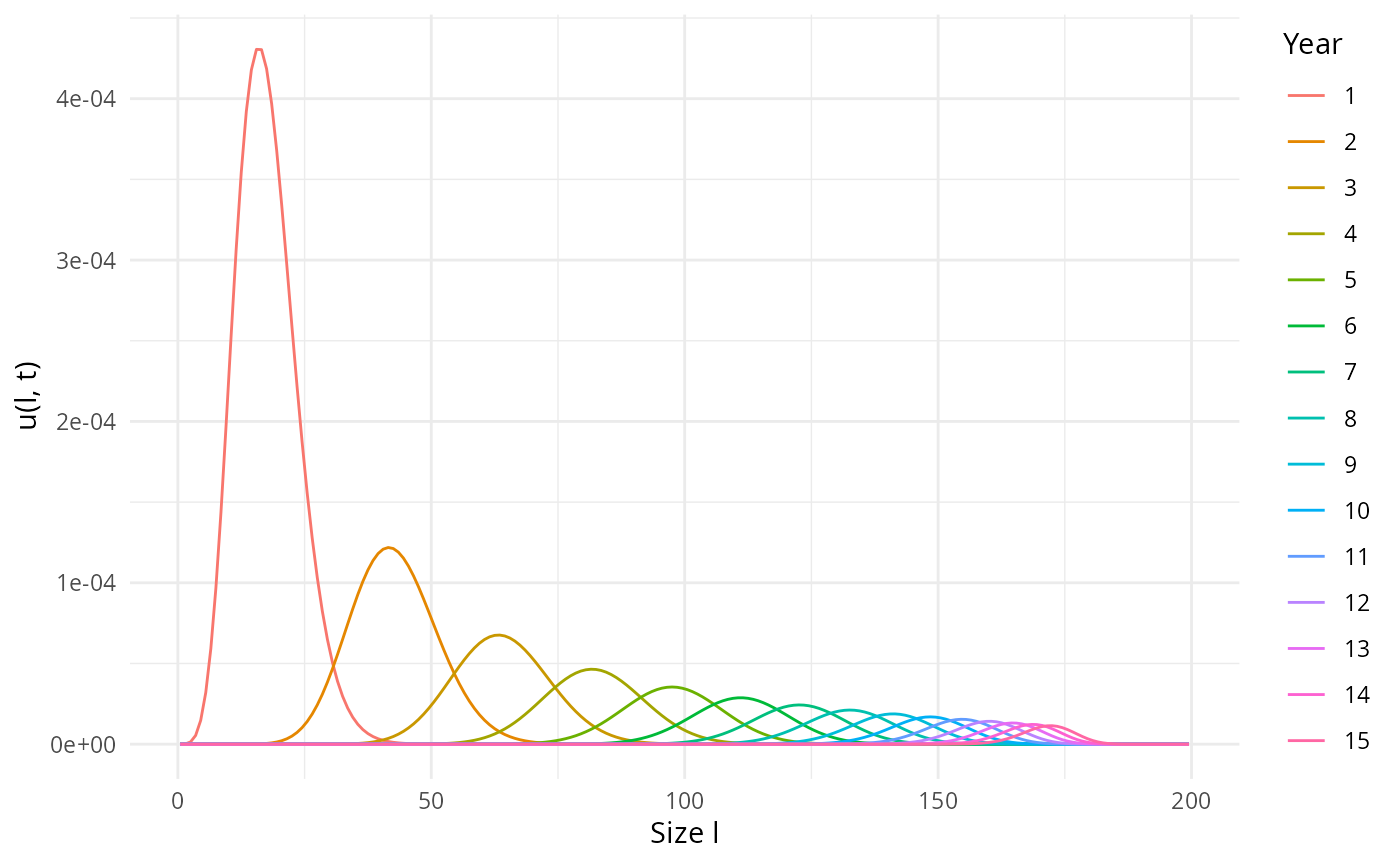
plotDensity3D(u, l_min = 20)The solution including all yearly cohorts looks as follows:
u_periodic <- getPeriodicNumberDensity(pars, l_max = 200, t_max = 3)
plotDensity3D(u_periodic, l_min = 20)